Sequence Spotlight: Tackling RSV with molecular assays
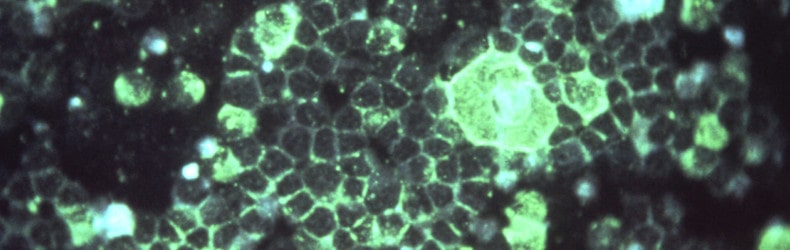
This summer, the Intensive Care Unit at Maimonides Children's Hospital in Brooklyn was overwhelmed by patients coughing and struggling to breathe – but the virus responsible wasn’t SARS-Cov-2.
What was causing the crisis – and similar outbreaks in the southern US, Switzerland, the UK, and Japan – was respiratory syncytial virus (RSV). RSV is a leading cause of acute respiratory infection that may be life-threatening to high-risk babies and vulnerable older people, and can cause serious complications including pneumonia, chronic obstructive pulmonary disease and heart failure. Every year, an estimated 177,000 older adults are hospitalised with RSV in the US, with 14,000 of them dying.
While they have the potential to be lethal, RSV outbreaks are also normally predictable, as they tend to take place in winter. Yet between July and August this year, American RSV infections were rising faster than even coronavirus cases. In fact, doctors believe that the atypical RSV surge was an indirect result of the coronavirus pandemic - because while lockdowns and hygiene measures may have suppressed the spread of SARS-Cov-2 and RSV last year, they also meant people were unable to build up immunity against them. Once restrictions were loosened, RSV simply found more people who were ready to infect.
Because RSV presents in a very similar mannner to other respiratory infections, we need to be able to test for it rapidly and accurately in the laboratory so that the right treatment can be given, as soon as possible. But, particularly in the case of RSV and other viral infections, rapid and accurate molecular assays can also help with the much more fundamental problem of antimicrobial resistance, by minimising the unnecessary use of antibiotics.
Using reference-grade RSV-A and RSV-B genomes, strains that are now available from ATCC’s Genome Portal, researchers based in Finland successfully developed a fast and accurate molecular assay for RSV using RT-SIBA. The developed assay demonstrated 100 per cent sensitivity and specificity, while also shortening the time needed to diagnose RSV from nasopharyngeal swab specimens from two hours to just 20 minutes.
ATCC’s role in combating RSV is just part of its ongoing commitment to better global health via the Genome Portal – our free-to-access database of more than 1,500 publicly-available bacteria, virus and other genomes and supporting metadata, matched to authenticated ATCC strains.
State-of-the-art quality control techniques employed by ATCC Genome Portal scientists use a hybrid assembly method that leverages highly accurate Illumina short-reads with the scaffolding ability of ultra-long Oxford Nanopore reads - generating a genome assembly more complete and accurate than could be possible with each individual technology.
ATCC provides scientists with the biomaterials, services and standards that move their research forward - and their Genome Portal makes it easy to access and analyse high-quality whole-genome sequencing data. Click here now to sign up free.
